Lab 2: Cluster analysis of sequential data¶
About the dataset¶
In this lab, we are going to use the built-in biofam data set from the TraMineR package. See more details here
This data consists information about the Family life states from the Swiss Household Panel biographical survey. 16 year-long family life sequences built from the retrospective biographical survey carried out by the Swiss Household Panel (SHP) in 2002.
A data frame with 2000 rows, 16 state variables, 1 id variable and 7 covariates and 2 weights variables.
The data set contains (in columns 10 to 25) sequences of family life states from age 15 to 30 (sequence length is 16) and a series of covariates. The sequences are a sample of 2000 sequences of those created from the SHP biographical survey. It includes only individuals who were at least 30 years old at the time of the survey. The biofam data set describes family life courses of 2000 individuals born between 1909 and 1972.
The states numbered from 0 to 7 are defined from the combination of five basic states, namely Living with parents (Parent), Left home (Left), Married (Marr), Having Children (Child), Divorced:
0 = “Parent”
1 = “Left”
2 = “Married”
3 = “Left+Marr”
4 = “Child”
5 = “Left+Child”
6 = “Left+Marr+Child”
7 = “Divorced”
Variable |
Label |
|---|---|
idhous |
ID |
sex |
sex |
birthy |
birth year |
nat102 |
nationality |
plingu02 |
interview language |
p02r01 |
confession or religion |
p02r04 |
participation in religious services: frequency |
cspfaj |
Swiss socio-professional category: Fathers job |
cspmoj |
Swiss socio-professional category: Mothers job |
a15 |
family status at age 15 |
… |
|
a30 |
family status at age 30 |
library(tidyverse)
library(TraMineR)
library(cluster)
data(biofam)
str(biofam)
── Attaching packages ──────────────────────────────────────────────────────────────────────────── tidyverse 1.3.1 ──
✔ ggplot2 3.3.5 ✔ purrr 0.3.4
✔ tibble 3.1.5 ✔ dplyr 1.0.7
✔ tidyr 1.1.3 ✔ stringr 1.4.0
✔ readr 2.0.1 ✔ forcats 0.5.1
Warning message:
“package ‘tibble’ was built under R version 4.1.1”
Warning message:
“package ‘readr’ was built under R version 4.1.1”
── Conflicts ─────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
Warning message:
“package ‘TraMineR’ was built under R version 4.1.1”
TraMineR stable version 2.2-3 (Built: 2022-01-26)
Website: http://traminer.unige.ch
Please type 'citation("TraMineR")' for citation information.
Warning message:
“package ‘cluster’ was built under R version 4.1.1”
'data.frame': 2000 obs. of 27 variables:
$ idhous : num 66891 28621 57711 17501 147701 ...
$ sex : Factor w/ 2 levels "man","woman": 1 1 2 1 1 1 1 1 1 2 ...
$ birthyr : num 1943 1935 1946 1918 1946 ...
$ nat_1_02: Factor w/ 200 levels "other error",..: 6 6 6 6 6 6 6 6 6 6 ...
$ plingu02: Factor w/ 3 levels "french","german",..: 2 2 1 2 2 3 2 1 1 2 ...
$ p02r01 : Factor w/ 13 levels "other error",..: 6 7 13 7 7 7 6 9 6 7 ...
$ p02r04 : Factor w/ 14 levels "other error",..: 9 13 7 13 7 6 7 14 9 13 ...
$ cspfaj : Factor w/ 12 levels "active occupied but not classified",..: 7 7 7 5 NA 12 NA 11 7 7 ...
$ cspmoj : Factor w/ 12 levels "active occupied but not classified",..: 7 NA 9 NA NA NA NA NA 7 NA ...
$ a15 : num 0 0 0 0 0 0 0 0 0 1 ...
$ a16 : num 0 1 0 0 0 0 0 0 0 1 ...
$ a17 : num 0 1 0 0 0 0 0 0 0 1 ...
$ a18 : num 0 1 0 0 0 0 0 0 0 1 ...
$ a19 : num 0 1 0 0 0 0 0 0 0 1 ...
$ a20 : num 0 1 0 1 1 0 0 0 0 1 ...
$ a21 : num 0 1 0 1 1 0 0 1 0 1 ...
$ a22 : num 0 1 1 1 1 0 0 1 0 1 ...
$ a23 : num 0 1 1 1 1 0 0 1 0 1 ...
$ a24 : num 3 1 1 1 1 0 2 1 0 6 ...
$ a25 : num 6 1 1 1 1 0 2 1 0 6 ...
$ a26 : num 6 3 1 1 1 0 2 3 6 6 ...
$ a27 : num 6 6 3 1 1 0 2 3 6 6 ...
$ a28 : num 6 6 6 1 6 0 2 3 6 6 ...
$ a29 : num 6 6 6 1 6 0 2 6 6 6 ...
$ a30 : num 6 6 6 1 6 0 2 6 6 6 ...
$ wp00tbgp: num 1053 855 575 1527 796 ...
$ wp00tbgs: num 0.935 0.759 0.51 1.356 0.707 ...
# state labels
bfstates <- c("Parent", "Left", "Married", "Left+Marr", "Child", "Left+Child", "Left+Marr+Child", "Divorced")
# define sequence object
biofam.seq <- seqdef(biofam, 10:25, states = bfstates, labels = bfstates)
[>] state coding:
[alphabet] [label] [long label]
1 0 Parent Parent
2 1 Left Left
3 2 Married Married
4 3 Left+Marr Left+Marr
5 4 Child Child
6 5 Left+Child Left+Child
7 6 Left+Marr+Child Left+Marr+Child
8 7 Divorced Divorced
[>] 2000 sequences in the data set
[>] min/max sequence length: 16/16
Q1. Create a normalized dissimilarity matrix using Longest Common Subsequences method¶
Store the dissimilarity matrix in biofam.seq.LCS
biofam.seq.LCS <- NULL
# BEGIN SOLUTION
biofam.seq.LCS <- seqdist(biofam.seq, method='LCS', norm = 'auto')
biofam.seq.LCS
# END SOLUTION
[>] 2000 sequences with 8 distinct states
[>] creating a 'sm' with a substitution cost of 2
[>] creating 8x8 substitution-cost matrix using 2 as constant value
[>] 537 distinct sequences
[>] min/max sequence lengths: 16/16
[>] computing distances using the LCS gmean normalized metric
[>] elapsed time: 0.214 secs
| 1167 | 514 | 1013 | 275 | 2580 | 773 | 1187 | 47 | 2091 | 1846 | ⋯ | 278 | 1980 | 787 | 1120 | 59 | 629 | 2297 | 775 | 2522 | 719 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1167 | 0.0000 | 0.6250 | 0.3125 | 0.6875 | 0.5000 | 0.4375 | 0.4375 | 0.4375 | 0.1250 | 0.6250 | ⋯ | 0.4375 | 0.6875 | 0.4375 | 0.3750 | 0.4375 | 0.2500 | 0.5625 | 0.4375 | 0.8125 | 0.2500 |
| 514 | 0.6250 | 0.0000 | 0.3750 | 0.3125 | 0.2500 | 0.9375 | 0.9375 | 0.4375 | 0.6875 | 0.1875 | ⋯ | 0.9375 | 0.3125 | 0.8125 | 0.8750 | 0.9375 | 0.5000 | 0.2500 | 0.9375 | 0.9375 | 0.6875 |
| 1013 | 0.3125 | 0.3750 | 0.0000 | 0.3750 | 0.1875 | 0.5625 | 0.5625 | 0.1250 | 0.3750 | 0.5000 | ⋯ | 0.5625 | 0.3750 | 0.4375 | 0.5000 | 0.5625 | 0.2500 | 0.3125 | 0.5625 | 0.8125 | 0.4375 |
| 275 | 0.6875 | 0.3125 | 0.3750 | 0.0000 | 0.1875 | 0.6875 | 0.6875 | 0.3750 | 0.6875 | 0.4375 | ⋯ | 0.6875 | 0.0000 | 0.5625 | 0.6875 | 0.6875 | 0.5000 | 0.5000 | 0.6875 | 0.8125 | 0.6875 |
| 2580 | 0.5000 | 0.2500 | 0.1875 | 0.1875 | 0.0000 | 0.6875 | 0.6875 | 0.2500 | 0.5000 | 0.3125 | ⋯ | 0.6875 | 0.1875 | 0.5625 | 0.6250 | 0.6875 | 0.3125 | 0.3125 | 0.6875 | 0.8125 | 0.5000 |
| 773 | 0.4375 | 0.9375 | 0.5625 | 0.6875 | 0.6875 | 0.0000 | 0.4375 | 0.6250 | 0.3125 | 1.0000 | ⋯ | 0.0000 | 0.6875 | 0.1250 | 0.0625 | 0.1875 | 0.6250 | 0.8750 | 0.0000 | 0.8125 | 0.6250 |
| 1187 | 0.4375 | 0.9375 | 0.5625 | 0.6875 | 0.6875 | 0.4375 | 0.0000 | 0.6250 | 0.4375 | 1.0000 | ⋯ | 0.4375 | 0.6875 | 0.4375 | 0.4375 | 0.2500 | 0.6250 | 0.8750 | 0.4375 | 0.3750 | 0.6250 |
| 47 | 0.4375 | 0.4375 | 0.1250 | 0.3750 | 0.2500 | 0.6250 | 0.6250 | 0.0000 | 0.5000 | 0.5625 | ⋯ | 0.6250 | 0.3750 | 0.5000 | 0.5625 | 0.6250 | 0.3125 | 0.2500 | 0.6250 | 0.8125 | 0.5000 |
| 2091 | 0.1250 | 0.6875 | 0.3750 | 0.6875 | 0.5000 | 0.3125 | 0.4375 | 0.5000 | 0.0000 | 0.6875 | ⋯ | 0.3125 | 0.6875 | 0.3125 | 0.2500 | 0.3125 | 0.3125 | 0.6250 | 0.3125 | 0.8125 | 0.3125 |
| 1846 | 0.6250 | 0.1875 | 0.5000 | 0.4375 | 0.3125 | 1.0000 | 1.0000 | 0.5625 | 0.6875 | 0.0000 | ⋯ | 1.0000 | 0.4375 | 0.8750 | 0.9375 | 1.0000 | 0.3750 | 0.3750 | 1.0000 | 1.0000 | 0.5625 |
| 1990 | 0.4375 | 0.8750 | 0.5000 | 0.6875 | 0.6875 | 0.5000 | 0.5000 | 0.4375 | 0.5000 | 1.0000 | ⋯ | 0.5000 | 0.6875 | 0.5000 | 0.5000 | 0.5000 | 0.6250 | 0.6250 | 0.5000 | 0.8125 | 0.6250 |
| 2088 | 0.4375 | 0.9375 | 0.5625 | 0.6875 | 0.6875 | 0.0000 | 0.4375 | 0.6250 | 0.3125 | 1.0000 | ⋯ | 0.0000 | 0.6875 | 0.1250 | 0.0625 | 0.1875 | 0.6250 | 0.8750 | 0.0000 | 0.8125 | 0.6250 |
| 867 | 0.2500 | 0.3750 | 0.1875 | 0.4375 | 0.2500 | 0.6875 | 0.6875 | 0.2500 | 0.3750 | 0.3750 | ⋯ | 0.6875 | 0.4375 | 0.5625 | 0.6250 | 0.6875 | 0.1250 | 0.3125 | 0.6875 | 0.8125 | 0.3125 |
| 1616 | 0.2500 | 0.6875 | 0.4375 | 0.6875 | 0.5000 | 0.6250 | 0.6250 | 0.5000 | 0.3125 | 0.5625 | ⋯ | 0.6250 | 0.6875 | 0.6250 | 0.5625 | 0.6250 | 0.1875 | 0.6250 | 0.6250 | 0.8125 | 0.0000 |
| 2136 | 0.4375 | 0.8750 | 0.5000 | 0.6875 | 0.6875 | 0.5000 | 0.5000 | 0.4375 | 0.5000 | 1.0000 | ⋯ | 0.5000 | 0.6875 | 0.5000 | 0.5000 | 0.5000 | 0.6250 | 0.6875 | 0.5000 | 0.8125 | 0.6250 |
| 2031 | 0.3750 | 0.6875 | 0.4375 | 0.6875 | 0.5000 | 0.6250 | 0.5000 | 0.5000 | 0.3750 | 0.7500 | ⋯ | 0.6250 | 0.6875 | 0.6250 | 0.5625 | 0.5000 | 0.3750 | 0.6250 | 0.6250 | 0.6875 | 0.3750 |
| 2459 | 0.4375 | 0.9375 | 0.5625 | 0.6875 | 0.6875 | 0.0000 | 0.4375 | 0.6250 | 0.3125 | 1.0000 | ⋯ | 0.0000 | 0.6875 | 0.1250 | 0.0625 | 0.1875 | 0.6250 | 0.8750 | 0.0000 | 0.8125 | 0.6250 |
| 222 | 0.4375 | 0.9375 | 0.5625 | 0.6875 | 0.6875 | 0.0000 | 0.4375 | 0.6250 | 0.3125 | 1.0000 | ⋯ | 0.0000 | 0.6875 | 0.1250 | 0.0625 | 0.1875 | 0.6250 | 0.8750 | 0.0000 | 0.8125 | 0.6250 |
| 2193 | 0.3750 | 0.8750 | 0.5000 | 0.6875 | 0.6875 | 0.1875 | 0.4375 | 0.4375 | 0.3125 | 1.0000 | ⋯ | 0.1875 | 0.6875 | 0.1875 | 0.1875 | 0.1875 | 0.6250 | 0.6875 | 0.1875 | 0.8125 | 0.6250 |
| 1571 | 0.3125 | 0.4375 | 0.3125 | 0.5625 | 0.3750 | 0.7500 | 0.7500 | 0.3750 | 0.4375 | 0.3750 | ⋯ | 0.7500 | 0.5625 | 0.6250 | 0.6875 | 0.7500 | 0.1250 | 0.3750 | 0.7500 | 0.8125 | 0.2500 |
| 2592 | 0.1250 | 0.6875 | 0.3750 | 0.6875 | 0.5000 | 0.5000 | 0.5000 | 0.5000 | 0.1875 | 0.5625 | ⋯ | 0.5000 | 0.6875 | 0.5000 | 0.4375 | 0.5000 | 0.1875 | 0.6250 | 0.5000 | 0.8125 | 0.1250 |
| 1989 | 0.3125 | 0.6250 | 0.5000 | 0.7500 | 0.5625 | 0.7500 | 0.7500 | 0.5000 | 0.4375 | 0.5625 | ⋯ | 0.7500 | 0.7500 | 0.7500 | 0.6875 | 0.7500 | 0.3125 | 0.5000 | 0.7500 | 0.8125 | 0.1250 |
| 1917 | 0.1250 | 0.6250 | 0.3125 | 0.6875 | 0.5000 | 0.5000 | 0.5000 | 0.3125 | 0.1875 | 0.6875 | ⋯ | 0.5000 | 0.6875 | 0.5000 | 0.4375 | 0.5000 | 0.3125 | 0.4375 | 0.5000 | 0.8125 | 0.3125 |
| 630 | 0.5625 | 0.5625 | 0.6250 | 0.8125 | 0.6250 | 0.9375 | 0.9375 | 0.6875 | 0.6250 | 0.4375 | ⋯ | 0.9375 | 0.8125 | 0.8125 | 0.8750 | 0.9375 | 0.3750 | 0.5625 | 0.9375 | 0.9375 | 0.3125 |
| 532 | 0.2500 | 0.6875 | 0.4375 | 0.6875 | 0.5000 | 0.6250 | 0.6250 | 0.5000 | 0.3125 | 0.5625 | ⋯ | 0.6250 | 0.6875 | 0.6250 | 0.5625 | 0.6250 | 0.1875 | 0.6250 | 0.6250 | 0.8125 | 0.0000 |
| 863 | 0.1875 | 0.6250 | 0.3750 | 0.6875 | 0.5000 | 0.6250 | 0.6250 | 0.3750 | 0.3125 | 0.5625 | ⋯ | 0.6250 | 0.6875 | 0.6250 | 0.5625 | 0.6250 | 0.1875 | 0.5000 | 0.6250 | 0.8125 | 0.1250 |
| 1102 | 0.5625 | 0.4375 | 0.5000 | 0.6875 | 0.5000 | 0.9375 | 0.9375 | 0.5625 | 0.6250 | 0.3125 | ⋯ | 0.9375 | 0.6875 | 0.8125 | 0.8750 | 0.9375 | 0.3125 | 0.4375 | 0.9375 | 0.9375 | 0.3125 |
| 1454 | 0.4375 | 0.9375 | 0.5625 | 0.6875 | 0.6875 | 0.0000 | 0.4375 | 0.6250 | 0.3125 | 1.0000 | ⋯ | 0.0000 | 0.6875 | 0.1250 | 0.0625 | 0.1875 | 0.6250 | 0.8750 | 0.0000 | 0.8125 | 0.6250 |
| 1174 | 0.4375 | 0.2500 | 0.1250 | 0.2500 | 0.0625 | 0.6875 | 0.6875 | 0.1875 | 0.5000 | 0.3750 | ⋯ | 0.6875 | 0.2500 | 0.5625 | 0.6250 | 0.6875 | 0.3125 | 0.2500 | 0.6875 | 0.8125 | 0.5000 |
| 227 | 0.3125 | 0.6875 | 0.5000 | 0.6875 | 0.5000 | 0.6875 | 0.6875 | 0.5625 | 0.3750 | 0.5625 | ⋯ | 0.6875 | 0.6875 | 0.6875 | 0.6250 | 0.6875 | 0.2500 | 0.6250 | 0.6875 | 0.8125 | 0.0625 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋱ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| 81 | 0.5625 | 0.3750 | 0.2500 | 0.1250 | 0.1875 | 0.5625 | 0.5625 | 0.3125 | 0.5625 | 0.4375 | ⋯ | 0.5625 | 0.1250 | 0.4375 | 0.5625 | 0.5625 | 0.4375 | 0.5000 | 0.5625 | 0.8125 | 0.6250 |
| 1805 | 0.1875 | 0.6875 | 0.3750 | 0.6875 | 0.5000 | 0.3125 | 0.4375 | 0.5000 | 0.0625 | 0.7500 | ⋯ | 0.3125 | 0.6875 | 0.3125 | 0.2500 | 0.3125 | 0.3750 | 0.6250 | 0.3125 | 0.8125 | 0.3750 |
| 789 | 0.9375 | 0.3125 | 0.6250 | 0.2500 | 0.4375 | 0.9375 | 0.9375 | 0.6250 | 0.9375 | 0.4375 | ⋯ | 0.9375 | 0.2500 | 0.8125 | 0.9375 | 0.9375 | 0.7500 | 0.5625 | 0.9375 | 0.9375 | 0.9375 |
| 2361 | 0.5625 | 0.9375 | 0.5625 | 0.6875 | 0.6875 | 0.5625 | 0.1250 | 0.6250 | 0.5625 | 1.0000 | ⋯ | 0.5625 | 0.6875 | 0.5625 | 0.5625 | 0.3750 | 0.6250 | 0.8750 | 0.5625 | 0.2500 | 0.6250 |
| 56 | 0.0625 | 0.6250 | 0.3125 | 0.6875 | 0.5000 | 0.5000 | 0.5000 | 0.3750 | 0.1875 | 0.6250 | ⋯ | 0.5000 | 0.6875 | 0.5000 | 0.4375 | 0.5000 | 0.2500 | 0.5000 | 0.5000 | 0.8125 | 0.2500 |
| 645 | 0.1875 | 0.5625 | 0.2500 | 0.5625 | 0.3750 | 0.5625 | 0.5625 | 0.3750 | 0.2500 | 0.4375 | ⋯ | 0.5625 | 0.5625 | 0.4375 | 0.5000 | 0.5625 | 0.0625 | 0.5000 | 0.5625 | 0.8125 | 0.1875 |
| 1721 | 0.1875 | 0.5625 | 0.1875 | 0.5625 | 0.3750 | 0.4375 | 0.4375 | 0.2500 | 0.2500 | 0.6875 | ⋯ | 0.4375 | 0.5625 | 0.3125 | 0.3750 | 0.4375 | 0.3125 | 0.4375 | 0.4375 | 0.8125 | 0.4375 |
| 1419 | 0.4375 | 0.9375 | 0.5625 | 0.6875 | 0.6875 | 0.1250 | 0.3125 | 0.6250 | 0.3125 | 1.0000 | ⋯ | 0.1250 | 0.6875 | 0.1250 | 0.1250 | 0.0625 | 0.6250 | 0.8750 | 0.1250 | 0.6875 | 0.6250 |
| 1207 | 0.6875 | 0.3125 | 0.3750 | 0.0000 | 0.1875 | 0.6875 | 0.6875 | 0.3750 | 0.6875 | 0.4375 | ⋯ | 0.6875 | 0.0000 | 0.5625 | 0.6875 | 0.6875 | 0.5000 | 0.5000 | 0.6875 | 0.8125 | 0.6875 |
| 259 | 0.4375 | 0.9375 | 0.5625 | 0.6875 | 0.6875 | 0.4375 | 0.0000 | 0.6250 | 0.4375 | 1.0000 | ⋯ | 0.4375 | 0.6875 | 0.4375 | 0.4375 | 0.2500 | 0.6250 | 0.8750 | 0.4375 | 0.3750 | 0.6250 |
| 2413 | 0.4375 | 0.3125 | 0.3125 | 0.5625 | 0.3750 | 0.8750 | 0.8750 | 0.2500 | 0.5625 | 0.3125 | ⋯ | 0.8750 | 0.5625 | 0.7500 | 0.8125 | 0.8750 | 0.3125 | 0.1250 | 0.8750 | 0.8750 | 0.5000 |
| 2090 | 0.6875 | 0.3125 | 0.3750 | 0.0000 | 0.1875 | 0.6875 | 0.6875 | 0.3750 | 0.6875 | 0.4375 | ⋯ | 0.6875 | 0.0000 | 0.5625 | 0.6875 | 0.6875 | 0.5000 | 0.5000 | 0.6875 | 0.8125 | 0.6875 |
| 1337 | 0.0625 | 0.6250 | 0.3125 | 0.6875 | 0.5000 | 0.4375 | 0.4375 | 0.3750 | 0.1250 | 0.6875 | ⋯ | 0.4375 | 0.6875 | 0.4375 | 0.3750 | 0.4375 | 0.3125 | 0.5000 | 0.4375 | 0.8125 | 0.3125 |
| 1826 | 0.2500 | 0.5000 | 0.3125 | 0.5625 | 0.3750 | 0.6875 | 0.6875 | 0.3125 | 0.3750 | 0.4375 | ⋯ | 0.6875 | 0.5625 | 0.5625 | 0.6250 | 0.6875 | 0.1250 | 0.3750 | 0.6875 | 0.8125 | 0.2500 |
| 2503 | 0.3750 | 0.8750 | 0.5000 | 0.6875 | 0.6875 | 0.3750 | 0.4375 | 0.4375 | 0.3750 | 1.0000 | ⋯ | 0.3750 | 0.6875 | 0.3750 | 0.3750 | 0.3750 | 0.6250 | 0.6250 | 0.3750 | 0.8125 | 0.6250 |
| 106 | 0.0625 | 0.6875 | 0.3750 | 0.6875 | 0.5000 | 0.4375 | 0.4375 | 0.5000 | 0.1250 | 0.5625 | ⋯ | 0.4375 | 0.6875 | 0.4375 | 0.3750 | 0.4375 | 0.1875 | 0.6250 | 0.4375 | 0.8125 | 0.1875 |
| 1181 | 0.3750 | 0.5000 | 0.1875 | 0.3125 | 0.3125 | 0.4375 | 0.4375 | 0.2500 | 0.4375 | 0.6250 | ⋯ | 0.4375 | 0.3125 | 0.3125 | 0.4375 | 0.4375 | 0.4375 | 0.4375 | 0.4375 | 0.8125 | 0.6250 |
| 1848 | 0.4375 | 0.8750 | 0.5000 | 0.6875 | 0.6875 | 0.5000 | 0.5000 | 0.4375 | 0.5000 | 1.0000 | ⋯ | 0.5000 | 0.6875 | 0.5000 | 0.5000 | 0.5000 | 0.6250 | 0.6250 | 0.5000 | 0.8125 | 0.6250 |
| 2203 | 0.6250 | 0.3125 | 0.3125 | 0.0625 | 0.1875 | 0.6250 | 0.6250 | 0.3125 | 0.6250 | 0.4375 | ⋯ | 0.6250 | 0.0625 | 0.5000 | 0.6250 | 0.6250 | 0.4375 | 0.5000 | 0.6250 | 0.8125 | 0.6250 |
| 1745 | 0.6875 | 0.0625 | 0.4375 | 0.2500 | 0.2500 | 0.9375 | 0.9375 | 0.5000 | 0.6875 | 0.1875 | ⋯ | 0.9375 | 0.2500 | 0.8125 | 0.8750 | 0.9375 | 0.5000 | 0.3125 | 0.9375 | 0.9375 | 0.6875 |
| 278 | 0.4375 | 0.9375 | 0.5625 | 0.6875 | 0.6875 | 0.0000 | 0.4375 | 0.6250 | 0.3125 | 1.0000 | ⋯ | 0.0000 | 0.6875 | 0.1250 | 0.0625 | 0.1875 | 0.6250 | 0.8750 | 0.0000 | 0.8125 | 0.6250 |
| 1980 | 0.6875 | 0.3125 | 0.3750 | 0.0000 | 0.1875 | 0.6875 | 0.6875 | 0.3750 | 0.6875 | 0.4375 | ⋯ | 0.6875 | 0.0000 | 0.5625 | 0.6875 | 0.6875 | 0.5000 | 0.5000 | 0.6875 | 0.8125 | 0.6875 |
| 787 | 0.4375 | 0.8125 | 0.4375 | 0.5625 | 0.5625 | 0.1250 | 0.4375 | 0.5000 | 0.3125 | 0.8750 | ⋯ | 0.1250 | 0.5625 | 0.0000 | 0.1250 | 0.1875 | 0.5000 | 0.7500 | 0.1250 | 0.8125 | 0.6250 |
| 1120 | 0.3750 | 0.8750 | 0.5000 | 0.6875 | 0.6250 | 0.0625 | 0.4375 | 0.5625 | 0.2500 | 0.9375 | ⋯ | 0.0625 | 0.6875 | 0.1250 | 0.0000 | 0.1875 | 0.5625 | 0.8125 | 0.0625 | 0.8125 | 0.5625 |
| 59 | 0.4375 | 0.9375 | 0.5625 | 0.6875 | 0.6875 | 0.1875 | 0.2500 | 0.6250 | 0.3125 | 1.0000 | ⋯ | 0.1875 | 0.6875 | 0.1875 | 0.1875 | 0.0000 | 0.6250 | 0.8750 | 0.1875 | 0.6250 | 0.6250 |
| 629 | 0.2500 | 0.5000 | 0.2500 | 0.5000 | 0.3125 | 0.6250 | 0.6250 | 0.3125 | 0.3125 | 0.3750 | ⋯ | 0.6250 | 0.5000 | 0.5000 | 0.5625 | 0.6250 | 0.0000 | 0.4375 | 0.6250 | 0.8125 | 0.1875 |
| 2297 | 0.5625 | 0.2500 | 0.3125 | 0.5000 | 0.3125 | 0.8750 | 0.8750 | 0.2500 | 0.6250 | 0.3750 | ⋯ | 0.8750 | 0.5000 | 0.7500 | 0.8125 | 0.8750 | 0.4375 | 0.0000 | 0.8750 | 0.8750 | 0.6250 |
| 775 | 0.4375 | 0.9375 | 0.5625 | 0.6875 | 0.6875 | 0.0000 | 0.4375 | 0.6250 | 0.3125 | 1.0000 | ⋯ | 0.0000 | 0.6875 | 0.1250 | 0.0625 | 0.1875 | 0.6250 | 0.8750 | 0.0000 | 0.8125 | 0.6250 |
| 2522 | 0.8125 | 0.9375 | 0.8125 | 0.8125 | 0.8125 | 0.8125 | 0.3750 | 0.8125 | 0.8125 | 1.0000 | ⋯ | 0.8125 | 0.8125 | 0.8125 | 0.8125 | 0.6250 | 0.8125 | 0.8750 | 0.8125 | 0.0000 | 0.8125 |
| 719 | 0.2500 | 0.6875 | 0.4375 | 0.6875 | 0.5000 | 0.6250 | 0.6250 | 0.5000 | 0.3125 | 0.5625 | ⋯ | 0.6250 | 0.6875 | 0.6250 | 0.5625 | 0.6250 | 0.1875 | 0.6250 | 0.6250 | 0.8125 | 0.0000 |
Q2. Plot the pairs of sequences¶
Plot the top 5 sequences that are the most similar to sequence 1
Plot the top 5 sequences that are the least similar to sequence 1
most_sim <- head(which(biofam.seq.LCS==min(biofam.seq.LCS), arr.ind=T))
most_sim
least_sim <- head(which(biofam.seq.LCS==max(biofam.seq.LCS), arr.ind=T))
least_sim
| row | col | |
|---|---|---|
| 1167 | 1 | 1 |
| 1746 | 39 | 1 |
| 2577 | 284 | 1 |
| 478 | 767 | 1 |
| 480 | 805 | 1 |
| 2380 | 919 | 1 |
| row | col | |
|---|---|---|
| 2304 | 443 | 1 |
| 2395 | 602 | 1 |
| 909 | 872 | 1 |
| 821 | 1461 | 1 |
| 769 | 1546 | 1 |
| 1012 | 1723 | 1 |
# BEGIN SOLUTION
seqiplot(biofam.seq[c(1,most_sim[,1]),])
seqiplot(biofam.seq[c(1,least_sim[,1]),])
# END SOLUTION
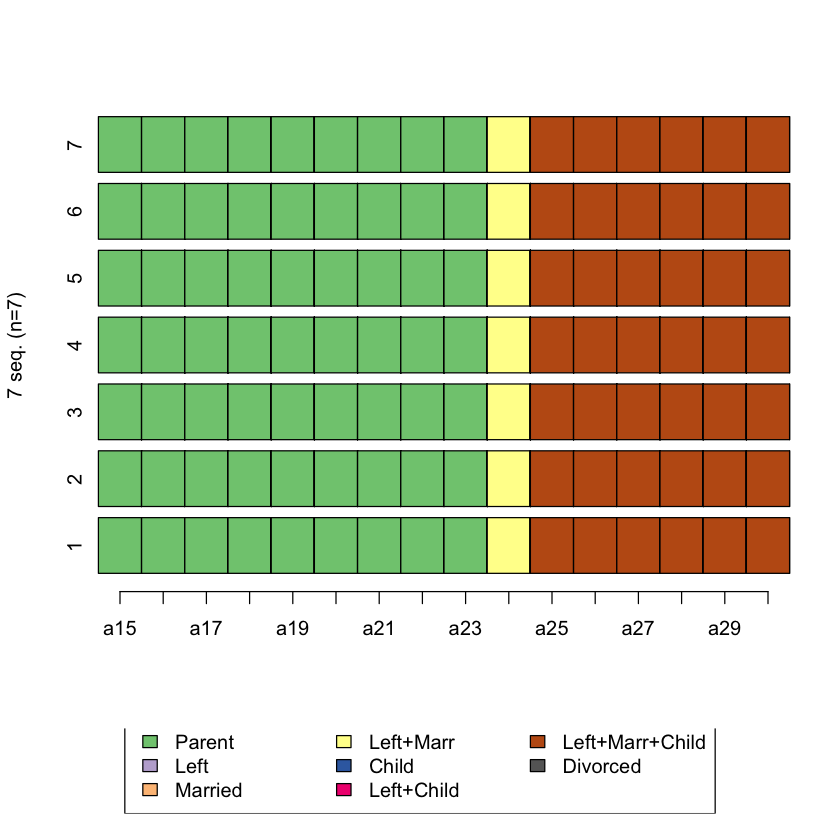
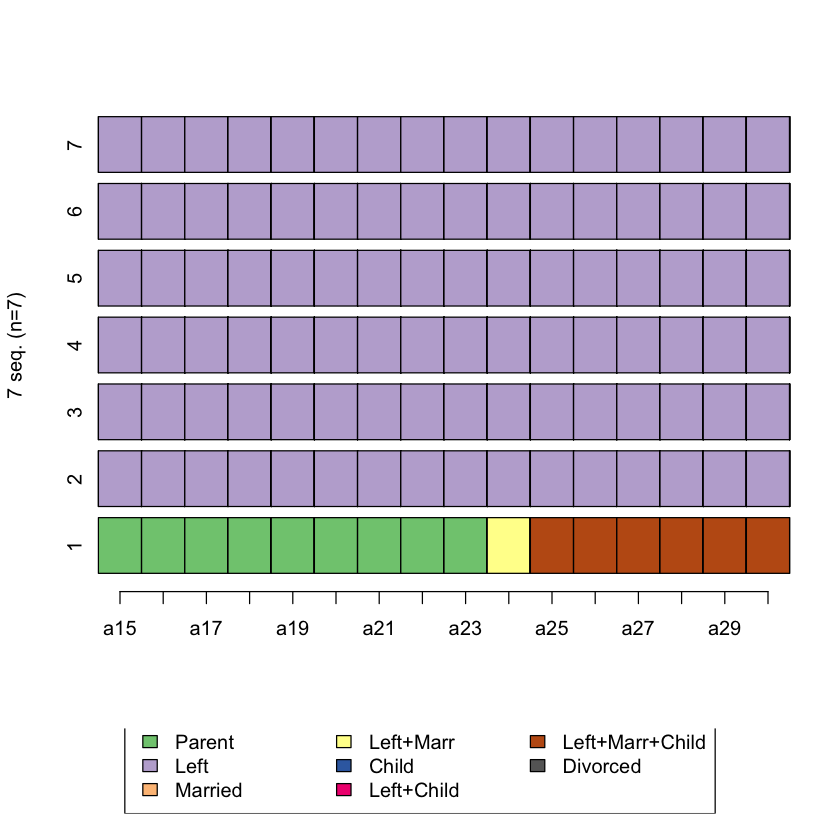
Q3. Create a dissimilarity matrix using optimal matching using transition rates as substitution cost matrix¶
biofam.seq.subcost <- NULL
biofam.seq.OM <- NULL
# BEGIN SOLUTION
biofam.seq.subcost <- seqcost(biofam.seq, method = "TRATE")
biofam.seq.OM <- seqdist(biofam.seq, method='OM', sm=biofam.seq.subcost$sm)
biofam.seq.OM
# END SOLUTION
[>] creating substitution-cost matrix using transition rates ...
[>] computing transition probabilities for states Parent/Left/Married/Left+Marr/Child/Left+Child/Left+Marr+Child/Divorced ...
[>] 2000 sequences with 8 distinct states
[>] checking 'sm' (size and triangle inequality)
[>] 537 distinct sequences
[>] min/max sequence lengths: 16/16
[>] computing distances using the OM metric
[>] elapsed time: 0.118 secs
| 1167 | 514 | 1013 | 275 | 2580 | 773 | 1187 | 47 | 2091 | 1846 | ⋯ | 278 | 1980 | 787 | 1120 | 59 | 629 | 2297 | 775 | 2522 | 719 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1167 | 0.000000 | 19.563325 | 9.890831 | 21.562411 | 15.630326 | 13.901396 | 13.92295 | 13.437363 | 3.957049 | 19.309299 | ⋯ | 13.901396 | 21.562411 | 13.878381 | 11.912527 | 13.901428 | 7.636805 | 17.241790 | 13.901396 | 25.83089 | 7.767166 |
| 514 | 19.563325 | 0.000000 | 11.727078 | 9.916578 | 7.890831 | 29.377814 | 29.50852 | 13.328194 | 21.254715 | 5.777920 | ⋯ | 29.377814 | 9.916578 | 25.563325 | 27.388945 | 29.377846 | 15.482360 | 7.695148 | 29.377814 | 29.83603 | 21.414446 |
| 1013 | 9.890831 | 11.727078 | 0.000000 | 11.739494 | 5.807409 | 17.661866 | 17.84715 | 3.800558 | 11.559582 | 15.582221 | ⋯ | 17.661866 | 11.739494 | 13.836247 | 15.672997 | 17.661899 | 7.755282 | 9.945416 | 17.661866 | 25.89494 | 13.676236 |
| 275 | 21.562411 | 9.916578 | 11.739494 | 0.000000 | 5.932086 | 21.399572 | 21.78166 | 11.649872 | 21.559303 | 13.954724 | ⋯ | 21.399572 | 0.000000 | 17.508741 | 21.431519 | 21.563325 | 15.786949 | 15.643672 | 21.399572 | 25.96931 | 21.719034 |
| 2580 | 15.630326 | 7.890831 | 5.807409 | 5.932086 | 0.000000 | 21.529933 | 21.74830 | 7.579129 | 15.627218 | 9.945416 | ⋯ | 21.529933 | 5.932086 | 17.672494 | 19.541064 | 21.529966 | 9.854863 | 9.833155 | 21.529933 | 25.93595 | 15.786949 |
| 773 | 13.901396 | 29.377814 | 17.661866 | 21.399572 | 21.529933 | 0.000000 | 13.89259 | 19.609357 | 9.944347 | 31.430826 | ⋯ | 0.000000 | 21.399572 | 3.890831 | 1.988869 | 5.953968 | 19.758332 | 27.500692 | 0.000000 | 25.80053 | 19.888693 |
| 1187 | 13.922955 | 29.508520 | 17.847146 | 21.781663 | 21.748303 | 13.892592 | 0.00000 | 19.783030 | 13.913712 | 31.430901 | ⋯ | 13.892592 | 21.781663 | 13.923280 | 13.896816 | 7.938624 | 19.758407 | 27.565217 | 13.892592 | 11.90794 | 19.888768 |
| 47 | 13.437363 | 13.328194 | 3.800558 | 11.649872 | 7.579129 | 19.609357 | 19.78303 | 0.000000 | 15.128752 | 17.183336 | ⋯ | 19.609357 | 11.649872 | 15.836247 | 17.620488 | 19.630873 | 9.356397 | 7.945416 | 19.609357 | 25.90075 | 15.288483 |
| 2091 | 3.957049 | 21.254715 | 11.559582 | 21.559303 | 15.627218 | 9.944347 | 13.91371 | 15.128752 | 0.000000 | 21.486480 | ⋯ | 9.944347 | 21.559303 | 9.921332 | 7.955477 | 9.944379 | 9.813986 | 19.377592 | 9.944347 | 25.82165 | 9.944347 |
| 1846 | 19.309299 | 5.777920 | 15.582221 | 13.954724 | 9.945416 | 31.430826 | 31.43090 | 17.183336 | 21.486480 | 0.000000 | ⋯ | 31.430826 | 13.954724 | 27.617910 | 29.441957 | 31.430859 | 11.672494 | 11.209083 | 31.430826 | 31.75841 | 17.604580 |
| 1990 | 12.771527 | 26.569874 | 15.013399 | 21.168867 | 20.820808 | 15.745441 | 15.89590 | 13.241680 | 14.907330 | 30.083808 | ⋯ | 15.745441 | 21.168867 | 15.642236 | 15.577819 | 15.809924 | 18.411314 | 18.874726 | 15.745441 | 25.84067 | 18.382202 |
| 2088 | 13.901396 | 29.377814 | 17.661866 | 21.399572 | 21.529933 | 0.000000 | 13.89259 | 19.609357 | 9.944347 | 31.430826 | ⋯ | 0.000000 | 21.399572 | 3.890831 | 1.988869 | 5.953968 | 19.758332 | 27.500692 | 0.000000 | 25.80053 | 19.888693 |
| 867 | 7.781663 | 11.781663 | 6.000000 | 13.780749 | 7.848663 | 21.683059 | 21.70462 | 7.601116 | 11.738712 | 11.527636 | ⋯ | 21.683059 | 13.780749 | 17.890831 | 19.694189 | 21.683091 | 3.745974 | 9.491947 | 21.683059 | 25.89227 | 9.678059 |
| 1616 | 7.767166 | 21.414446 | 13.676236 | 21.719034 | 15.786949 | 19.888693 | 19.88877 | 15.288483 | 9.944347 | 17.604580 | ⋯ | 19.888693 | 21.719034 | 19.865678 | 17.899824 | 19.888725 | 5.932086 | 18.938618 | 19.888693 | 25.84277 | 0.000000 |
| 2136 | 13.576843 | 27.206889 | 15.540463 | 21.585088 | 21.237921 | 15.904540 | 15.89987 | 13.878695 | 15.389537 | 30.940726 | ⋯ | 15.904540 | 21.585088 | 15.801335 | 15.736918 | 15.875946 | 19.268232 | 21.634623 | 15.904540 | 25.84465 | 19.349071 |
| 2031 | 11.957176 | 21.716753 | 13.785214 | 21.854150 | 15.922778 | 19.924789 | 15.94599 | 15.590790 | 11.963244 | 23.644186 | ⋯ | 19.924789 | 21.854150 | 19.901774 | 17.935920 | 15.988880 | 11.971692 | 19.761011 | 19.924789 | 21.87136 | 11.953487 |
| 2459 | 13.901396 | 29.377814 | 17.661866 | 21.399572 | 21.529933 | 0.000000 | 13.89259 | 19.609357 | 9.944347 | 31.430826 | ⋯ | 0.000000 | 21.399572 | 3.890831 | 1.988869 | 5.953968 | 19.758332 | 27.500692 | 0.000000 | 25.80053 | 19.888693 |
| 222 | 13.901396 | 29.377814 | 17.661866 | 21.399572 | 21.529933 | 0.000000 | 13.89259 | 19.609357 | 9.944347 | 31.430826 | ⋯ | 0.000000 | 21.399572 | 3.890831 | 1.988869 | 5.953968 | 19.758332 | 27.500692 | 0.000000 | 25.80053 | 19.888693 |
| 2193 | 11.601116 | 27.055272 | 15.328194 | 21.313058 | 20.964999 | 5.904540 | 13.90765 | 13.727078 | 9.379412 | 30.865892 | ⋯ | 5.904540 | 21.313058 | 5.801335 | 5.736918 | 5.969023 | 19.193398 | 21.640674 | 5.904540 | 25.81558 | 19.323759 |
| 1571 | 9.836247 | 13.836247 | 10.000000 | 17.758111 | 11.826025 | 23.715382 | 23.71546 | 11.601116 | 13.771035 | 11.675602 | ⋯ | 23.715382 | 17.758111 | 19.945416 | 21.726512 | 23.715414 | 4.000000 | 11.292505 | 23.715382 | 25.88537 | 7.668751 |
| 2592 | 3.789427 | 21.350553 | 11.655421 | 21.655142 | 15.723056 | 15.910955 | 15.91103 | 15.224591 | 5.966608 | 17.540687 | ⋯ | 15.910955 | 21.655142 | 15.887940 | 13.922085 | 15.910987 | 5.868193 | 18.874726 | 15.910955 | 25.83432 | 3.977739 |
| 1989 | 9.968180 | 19.752825 | 15.916578 | 23.629412 | 17.697326 | 23.825053 | 23.82513 | 15.800558 | 13.880707 | 17.546903 | ⋯ | 23.825053 | 23.629412 | 23.802039 | 21.836184 | 23.825086 | 9.810517 | 15.491947 | 23.825053 | 25.85281 | 3.936360 |
| 1917 | 3.768738 | 19.451065 | 9.778571 | 21.472789 | 15.540703 | 15.848887 | 15.89193 | 9.890831 | 5.904540 | 21.081019 | ⋯ | 15.848887 | 21.472789 | 15.801335 | 13.860018 | 15.848919 | 9.408525 | 13.473052 | 15.848887 | 25.83670 | 9.379412 |
| 630 | 17.624605 | 17.619453 | 19.732643 | 25.796257 | 19.864171 | 29.746133 | 29.74621 | 21.333759 | 19.801786 | 13.809587 | ⋯ | 29.746133 | 25.796257 | 26.000000 | 27.757263 | 29.746165 | 11.977362 | 17.111679 | 29.746133 | 29.74627 | 9.857439 |
| 532 | 7.767166 | 21.414446 | 13.676236 | 21.719034 | 15.786949 | 19.888693 | 19.88877 | 15.288483 | 9.944347 | 17.604580 | ⋯ | 19.888693 | 21.719034 | 19.865678 | 17.899824 | 19.888725 | 5.932086 | 18.938618 | 19.888693 | 25.84277 | 0.000000 |
| 863 | 5.968180 | 19.643656 | 11.916578 | 21.597466 | 15.665380 | 19.847315 | 19.84739 | 11.800558 | 9.902968 | 17.483011 | ⋯ | 19.847315 | 21.597466 | 19.824300 | 17.858445 | 19.847347 | 5.810517 | 15.382778 | 19.847315 | 25.84436 | 3.601116 |
| 1102 | 17.537698 | 13.664729 | 15.777920 | 21.841533 | 15.909447 | 29.659225 | 29.65930 | 17.379035 | 19.714879 | 9.854863 | ⋯ | 29.659225 | 21.841533 | 25.890831 | 27.670356 | 29.659257 | 9.945416 | 13.156955 | 29.659225 | 29.76851 | 9.770532 |
| 1454 | 13.901396 | 29.377814 | 17.661866 | 21.399572 | 21.529933 | 0.000000 | 13.89259 | 19.609357 | 9.944347 | 31.430826 | ⋯ | 0.000000 | 21.399572 | 3.890831 | 1.988869 | 5.953968 | 19.758332 | 27.500692 | 0.000000 | 25.80053 | 19.888693 |
| 1174 | 13.781663 | 7.836247 | 3.890831 | 7.848663 | 1.916578 | 21.552698 | 21.73798 | 5.662551 | 15.450414 | 11.691389 | ⋯ | 21.552698 | 7.848663 | 17.727078 | 19.563828 | 21.552730 | 9.678059 | 7.916578 | 21.552698 | 25.92563 | 15.610145 |
| 227 | 9.756035 | 21.446392 | 15.665106 | 21.750980 | 15.818895 | 21.877563 | 21.87764 | 17.277352 | 11.933216 | 17.636526 | ⋯ | 21.877563 | 21.750980 | 21.854548 | 19.888693 | 21.877595 | 7.920955 | 18.970564 | 21.877563 | 25.84699 | 1.988869 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋱ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| 81 | 17.671580 | 11.945416 | 7.848663 | 3.890831 | 5.977362 | 17.508741 | 17.890831 | 9.727094 | 17.668472 | 14.000000 | ⋯ | 17.508741 | 3.890831 | 13.617910 | 17.540687 | 17.672494 | 13.864171 | 15.749733 | 17.508741 | 25.938624 | 19.785126 |
| 1805 | 5.838994 | 21.454157 | 11.578299 | 21.578020 | 15.645934 | 9.954408 | 13.924832 | 15.328194 | 1.881944 | 23.368424 | ⋯ | 9.954408 | 21.578020 | 9.931393 | 7.965539 | 9.954440 | 11.695930 | 19.577034 | 9.954408 | 25.832768 | 11.826291 |
| 789 | 29.344074 | 9.826025 | 19.521157 | 7.781663 | 13.713748 | 29.181235 | 29.563325 | 19.431535 | 29.340966 | 13.864171 | ⋯ | 29.181235 | 7.781663 | 25.290404 | 29.213181 | 29.344988 | 23.568611 | 17.521173 | 29.181235 | 29.890831 | 29.500697 |
| 2361 | 17.892267 | 29.617689 | 17.956315 | 21.890831 | 21.857472 | 17.861904 | 3.969312 | 19.892199 | 17.883024 | 31.540070 | ⋯ | 17.861904 | 21.890831 | 17.892592 | 17.866128 | 11.907936 | 19.867576 | 27.641296 | 17.861904 | 7.938624 | 19.888790 |
| 56 | 1.968180 | 19.534487 | 9.861993 | 21.533573 | 15.601488 | 15.869576 | 15.891135 | 11.691389 | 5.925230 | 19.280461 | ⋯ | 15.869576 | 21.533573 | 15.833155 | 13.880707 | 15.869608 | 7.607967 | 15.273610 | 15.869576 | 25.835909 | 7.578854 |
| 645 | 5.691389 | 17.427776 | 7.732643 | 17.732364 | 11.800279 | 17.812917 | 17.812992 | 11.301813 | 7.868570 | 13.617910 | ⋯ | 17.812917 | 17.732364 | 14.000000 | 15.824047 | 17.812949 | 1.945416 | 15.183336 | 17.812917 | 25.860785 | 5.943593 |
| 1721 | 5.800558 | 17.363883 | 5.807409 | 17.546903 | 11.614818 | 13.793800 | 13.945989 | 7.636805 | 7.491947 | 21.219025 | ⋯ | 13.793800 | 17.546903 | 10.000000 | 11.804930 | 13.793832 | 9.546531 | 13.781663 | 13.793800 | 25.853925 | 13.522447 |
| 1419 | 13.901418 | 29.377836 | 17.661888 | 21.508741 | 21.529955 | 3.969312 | 9.923280 | 19.609379 | 9.944368 | 31.430848 | ⋯ | 3.969312 | 21.508741 | 4.000000 | 3.973536 | 1.984656 | 19.758354 | 27.500713 | 3.969312 | 21.831216 | 19.888715 |
| 1207 | 21.562411 | 9.916578 | 11.739494 | 0.000000 | 5.932086 | 21.399572 | 21.781663 | 11.649872 | 21.559303 | 13.954724 | ⋯ | 21.399572 | 0.000000 | 17.508741 | 21.431519 | 21.563325 | 15.786949 | 15.643672 | 21.399572 | 25.969312 | 21.719034 |
| 259 | 13.922955 | 29.508520 | 17.847146 | 21.781663 | 21.748303 | 13.892592 | 0.000000 | 19.783030 | 13.913712 | 31.430901 | ⋯ | 13.892592 | 21.781663 | 13.923280 | 13.896816 | 7.938624 | 19.758407 | 27.565217 | 13.892592 | 11.907936 | 19.888768 |
| 2413 | 13.663439 | 9.778571 | 10.000000 | 17.681818 | 11.749733 | 27.564835 | 27.586393 | 8.000000 | 17.620488 | 9.524544 | ⋯ | 27.564835 | 17.681818 | 23.836247 | 25.575965 | 27.564867 | 9.691389 | 3.800558 | 27.564835 | 27.847719 | 15.160698 |
| 2090 | 21.562411 | 9.916578 | 11.739494 | 0.000000 | 5.932086 | 21.399572 | 21.781663 | 11.649872 | 21.559303 | 13.954724 | ⋯ | 21.399572 | 0.000000 | 17.508741 | 21.431519 | 21.563325 | 15.786949 | 15.643672 | 21.399572 | 25.969312 | 21.719034 |
| 1337 | 1.800558 | 19.479903 | 9.807409 | 21.501627 | 15.569541 | 13.880707 | 13.923749 | 11.636805 | 3.936360 | 21.109857 | ⋯ | 13.880707 | 21.501627 | 13.833155 | 11.891838 | 13.880739 | 9.437363 | 15.441232 | 13.880707 | 25.831685 | 9.567724 |
| 1826 | 7.859011 | 15.698240 | 9.916578 | 17.674688 | 11.742602 | 21.749277 | 21.749352 | 9.800558 | 11.804930 | 13.560233 | ⋯ | 21.749277 | 17.674688 | 18.000000 | 19.760407 | 21.749309 | 3.916578 | 11.437363 | 21.749277 | 25.870821 | 7.523893 |
| 2503 | 11.002789 | 26.627550 | 15.071076 | 21.226544 | 20.878485 | 11.809081 | 13.922702 | 13.299356 | 10.970970 | 30.300958 | ⋯ | 11.809081 | 21.226544 | 11.705875 | 11.641458 | 11.873563 | 18.628464 | 19.273610 | 11.809081 | 25.830638 | 18.758825 |
| 106 | 1.800558 | 21.318607 | 11.623475 | 21.623196 | 15.691110 | 13.922085 | 13.922160 | 15.192644 | 3.977739 | 17.508741 | ⋯ | 13.922085 | 21.623196 | 13.899070 | 11.933216 | 13.922117 | 5.836247 | 19.042348 | 13.922085 | 25.830097 | 5.966608 |
| 1181 | 12.000000 | 15.781663 | 6.000000 | 9.698240 | 9.691389 | 13.640674 | 13.989674 | 7.916578 | 13.600837 | 19.636805 | ⋯ | 13.640674 | 9.698240 | 9.781663 | 13.473052 | 13.771337 | 13.732643 | 14.000000 | 13.640674 | 25.897610 | 19.631337 |
| 1848 | 12.771527 | 26.569874 | 15.013399 | 21.168867 | 20.820808 | 15.745441 | 15.895901 | 13.241680 | 14.907330 | 30.083808 | ⋯ | 15.745441 | 21.168867 | 15.642236 | 15.577819 | 15.809924 | 18.411314 | 18.874726 | 15.745441 | 25.840675 | 18.382202 |
| 2203 | 19.616996 | 10.000000 | 9.794079 | 1.945416 | 5.954724 | 19.454157 | 19.836247 | 9.704456 | 19.613888 | 13.977362 | ⋯ | 19.454157 | 1.945416 | 15.563325 | 19.486103 | 19.617910 | 13.841533 | 15.666310 | 19.454157 | 25.953968 | 19.773619 |
| 1745 | 21.434627 | 1.916578 | 13.473052 | 8.000000 | 7.836247 | 29.355050 | 29.518846 | 15.244772 | 21.431519 | 5.954724 | ⋯ | 29.355050 | 8.000000 | 25.508741 | 27.366180 | 29.355082 | 15.659164 | 9.611726 | 29.355050 | 29.846352 | 21.591249 |
| 278 | 13.901396 | 29.377814 | 17.661866 | 21.399572 | 21.529933 | 0.000000 | 13.892592 | 19.609357 | 9.944347 | 31.430826 | ⋯ | 0.000000 | 21.399572 | 3.890831 | 1.988869 | 5.953968 | 19.758332 | 27.500692 | 0.000000 | 25.800528 | 19.888693 |
| 1980 | 21.562411 | 9.916578 | 11.739494 | 0.000000 | 5.932086 | 21.399572 | 21.781663 | 11.649872 | 21.559303 | 13.954724 | ⋯ | 21.399572 | 0.000000 | 17.508741 | 21.431519 | 21.563325 | 15.786949 | 15.643672 | 21.399572 | 25.969312 | 21.719034 |
| 787 | 13.878381 | 25.563325 | 13.836247 | 17.508741 | 17.672494 | 3.890831 | 13.923280 | 15.836247 | 9.921332 | 27.617910 | ⋯ | 3.890831 | 17.508741 | 0.000000 | 3.922778 | 5.984656 | 15.945416 | 23.781663 | 3.890831 | 25.831216 | 19.865678 |
| 1120 | 11.912527 | 27.388945 | 15.672997 | 21.431519 | 19.541064 | 1.988869 | 13.896816 | 17.620488 | 7.955477 | 29.441957 | ⋯ | 1.988869 | 21.431519 | 3.922778 | 0.000000 | 5.958192 | 17.769463 | 25.511822 | 1.988869 | 25.804752 | 17.899824 |
| 59 | 13.901428 | 29.377846 | 17.661899 | 21.563325 | 21.529966 | 5.953968 | 7.938624 | 19.630873 | 9.944379 | 31.430859 | ⋯ | 5.953968 | 21.563325 | 5.984656 | 5.958192 | 0.000000 | 19.758365 | 27.500724 | 5.953968 | 19.846560 | 19.888725 |
| 629 | 7.636805 | 15.482360 | 7.755282 | 15.786949 | 9.854863 | 19.758332 | 19.758407 | 9.356397 | 9.813986 | 11.672494 | ⋯ | 19.758332 | 15.786949 | 15.945416 | 17.769463 | 19.758365 | 0.000000 | 13.237921 | 19.758332 | 25.876128 | 5.932086 |
| 2297 | 17.241790 | 7.695148 | 9.945416 | 15.643672 | 9.833155 | 27.500692 | 27.565217 | 7.945416 | 19.377592 | 11.209083 | ⋯ | 27.500692 | 15.643672 | 23.781663 | 25.511822 | 27.500724 | 13.237921 | 0.000000 | 27.500692 | 27.859633 | 18.938618 |
| 775 | 13.901396 | 29.377814 | 17.661866 | 21.399572 | 21.529933 | 0.000000 | 13.892592 | 19.609357 | 9.944347 | 31.430826 | ⋯ | 0.000000 | 21.399572 | 3.890831 | 1.988869 | 5.953968 | 19.758332 | 27.500692 | 0.000000 | 25.800528 | 19.888693 |
| 2522 | 25.830891 | 29.836026 | 25.894939 | 25.969312 | 25.935952 | 25.800528 | 11.907936 | 25.900751 | 25.821648 | 31.758407 | ⋯ | 25.800528 | 25.969312 | 25.831216 | 25.804752 | 19.846560 | 25.876128 | 27.859633 | 25.800528 | 0.000000 | 25.842769 |
| 719 | 7.767166 | 21.414446 | 13.676236 | 21.719034 | 15.786949 | 19.888693 | 19.888768 | 15.288483 | 9.944347 | 17.604580 | ⋯ | 19.888693 | 21.719034 | 19.865678 | 17.899824 | 19.888725 | 5.932086 | 18.938618 | 19.888693 | 25.842769 | 0.000000 |
?seqdist
Q4. Perform an agglomerative clustering using ward linkage method¶
You should use the dissimilarity matrix with optimal matching generated in Q3
Plot the dendogram
clusterward <- NULL
# BEGIN SOLUTION
clusterward <- agnes(biofam.seq.OM, diss = TRUE, method = "ward")
# Run this to generate the dendogram
plot(clusterward, which.plot=2)
# END SOLUTION
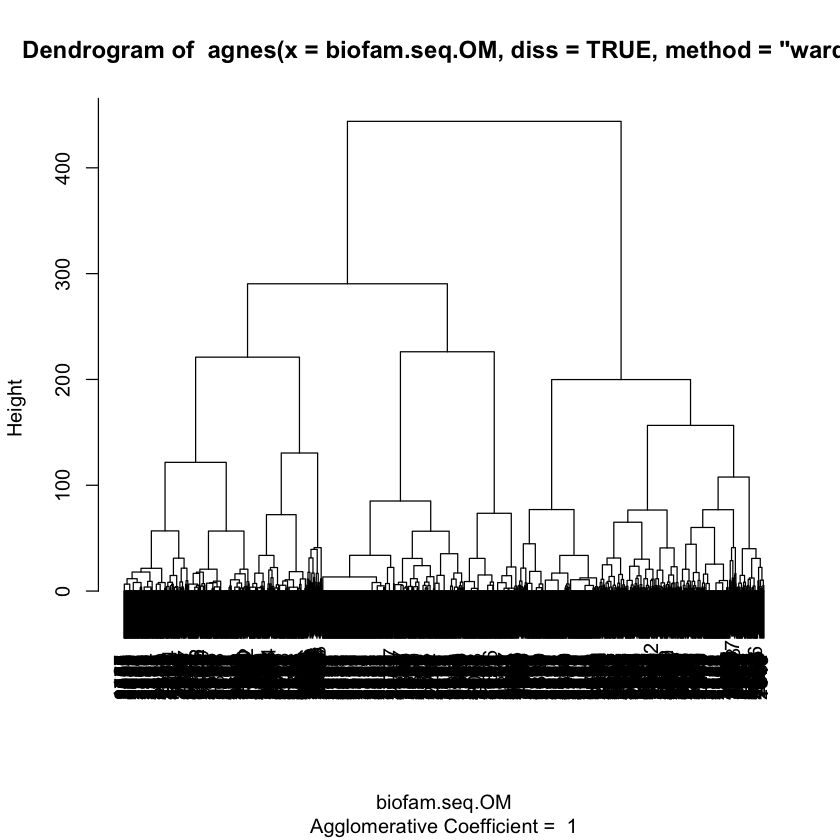
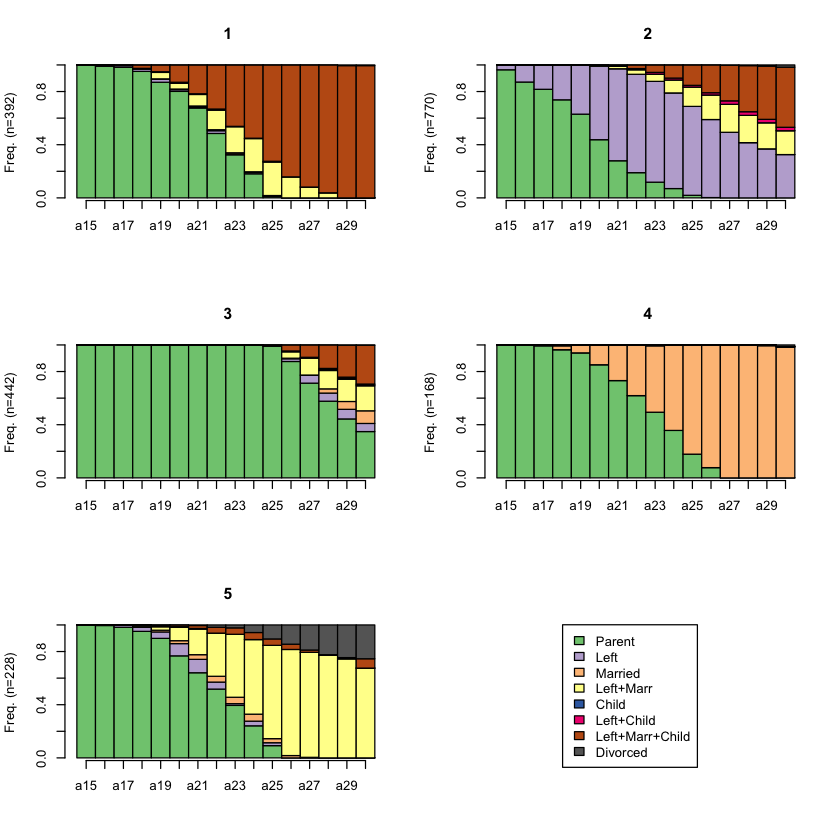
Q5: Select clusters¶
Cut the dendogram tree as appropriate using the
cutree()functionList the number of observations in each cluster
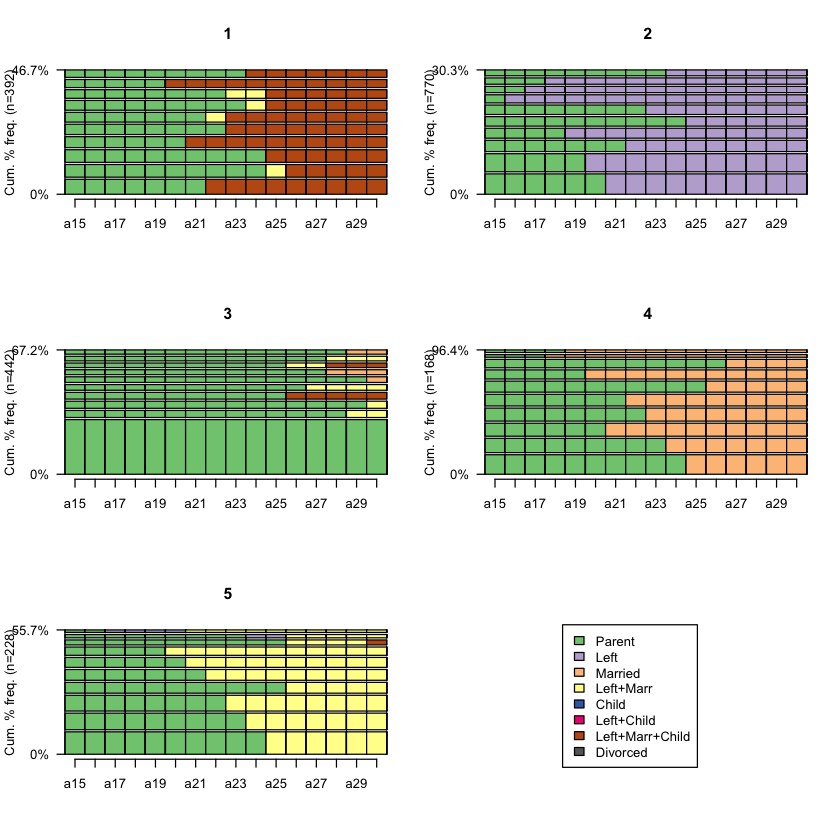
Plot the sequence frequency by cluster membership (hint:
seqfplot())Plot the state distribution by cluster membership (hint:
seqdplot())
# BEGIN SOLUTION
# cut the dendogram tree to generate two clusters
cluster5 <- cutree(clusterward, k = 5)
# check the number of observations in each cluster
table(cluster5)
# plot sequence frequency by cluster membership
seqfplot(biofam.seq, group = cluster5, pbarw = T)
# plot state distribution by cluster membership
seqdplot(biofam.seq, group = cluster5)
# END SOLUTION
s1 = Read (2) - Write (2)
s2 = Read(5) - Write (5)
cluster5
1 2 3 4 5
392 770 442 168 228
Error in Read(2): could not find function "Read"
Traceback:
?seqcost()
| seqcost {TraMineR} | R Documentation |
Generate substitution and indel costs
Description
The function seqcost proposes different ways to generate substitution costs
(supposed to represent state dissimilarities) and possibly indel costs. Proposed methods are:
"CONSTANT" (same cost for all substitutions), "TRATE" (derived from the observed transition rates), "FUTURE" (Chi-squared distance between conditional state distributions lag positions ahead), "FEATURES" (Gower distance between state features), "INDELS", "INDELSLOG" (based on estimated indel costs).
The substitution-cost matrix is intended to serve as sm argument in the seqdist function that computes distances between sequences. seqsubm is an alias that returns only the substitution cost matrix, i.e., no indel.
Usage
seqcost(seqdata, method, cval = NULL, with.missing = FALSE, miss.cost = NULL, time.varying = FALSE, weighted = TRUE, transition = "both", lag = 1, miss.cost.fixed = NULL, state.features = NULL, feature.weights = NULL, feature.type = list(), proximities = FALSE) seqsubm(...)
Arguments
seqdata |
A sequence object as returned by the seqdef function. |
method |
String. How to generate the costs. One of |
cval |
Scalar. For method |
with.missing |
Logical. Should an additional entry be added in the matrix for the missing states?
If |
miss.cost |
Scalar or vector. Cost for substituting the missing state. Default is |
miss.cost.fixed |
Logical. Should the substitution cost for missing be set as the |
time.varying |
Logical. If |
weighted |
Logical. Should weights in |
transition |
String. Only used if |
lag |
Integer. For methods |
state.features |
Data frame with features values for each state. |
feature.weights |
Vector of feature weights with length equal to the number of columns of |
feature.type |
List of feature types. See |
proximities |
Logical: should state proximities be returned instead of substitution costs? |
... |
Arguments passed to |
Details
The substitution-cost matrix has dimension ns*ns, where ns is the number of states in the alphabet of the sequence object. The element (i,j) of the matrix is the cost of substituting state i with state j. It represents the dissimilarity between the states i and j. The indel cost of the cost of inserting or deleting a state.
With method CONSTANT, the substitution costs are all set equal to the cval value, the default value being 2.
With method TRATE
(transition rates), the transition probabilities between all pairs of
states is first computed (using the seqtrate function). Then, the
substitution cost between states i and j is obtained with
the formula
SC(i,j) = cval - P(i|j) -P(j|i)
where P(i|j) is the probability of transition from state j to
i lag positions ahead. Default cval value is 2. When time.varying=TRUE and transition="both", the substitution cost at position t is set as
SC(i,j,t) = cval - P(i|j,t-1) -P(j|i,t-1) - P(i|j,t) - P(j|i,t)
where P(i|j,t-1) is the probability to transit from state j at t-1 to i at t. Here, the default cval value is 4.
With method FUTURE, the cost between i and j is the Chi-squared distance between the vector (d(alphabet | i)) of probabilities of transition from states i and
j to all the states in the alphabet lag positions ahead:
SC(i,j) = ChiDist(d(alphabet | i), d(alphabet | j))
With method FEATURES, each state is characterized by the variables state.features, and the cost between i and j is computed as the Gower distance between their vectors of state.features values.
With methods INDELS and INDELSLOG, values of indels are first derived from the state relative frequencies f_i. For INDELS, indel_i = 1/f_i is used, and for INDELSLOG, indel_i = log[2/(1 + f_i)].
Substitution costs are then set as SC(i,j) = indel_i + indel_j.
For all methods but INDELS and INDELSLOG, the indel is set as max(sm)/2 when time.varying=FALSE and as 1 otherwise.
Value
For seqcost, a list of two elements, indel and sm or prox:
indel |
The indel cost. Either a scalar or a vector of size ns. When |
sm |
The substitution-cost matrix (or array) when |
prox |
The state proximity matrix when |
sm and prox are, when time.varying = FALSE, a matrix of size ns * ns, where ns
is the number of states in the alphabet of the sequence object. When time.varying = TRUE, they are a three dimensional array of size ns * ns * L, where L is the maximum sequence length.
For seqsubm, only one element, the matrix (or array) sm.
Author(s)
Gilbert Ritschard and Matthias Studer (and Alexis Gabadinho for first version of seqsubm)
References
Gabadinho, A., G. Ritschard, N. S. Müller and M. Studer (2011). Analyzing and Visualizing State Sequences in R with TraMineR. Journal of Statistical Software 40(4), 1-37.
Gabadinho, A., G. Ritschard, M. Studer and N. S. Müller (2010). Mining Sequence Data in
R with the TraMineR package: A user's guide. Department of Econometrics and
Laboratory of Demography, University of Geneva.
Studer, M. & Ritschard, G. (2016), "What matters in differences between life trajectories: A comparative review of sequence dissimilarity measures", Journal of the Royal Statistical Society, Series A. 179(2), 481-511. doi: 10.1111/rssa.12125
Studer, M. and G. Ritschard (2014). "A Comparative Review of Sequence Dissimilarity Measures". LIVES Working Papers, 33. NCCR LIVES, Switzerland, 2014. doi: 10.12682/lives.2296-1658.2014.33
See Also
seqtrate, seqdef, seqdist.
Examples
## Defining a sequence object with columns 10 to 25 ## of a subset of the 'biofam' example data set. data(biofam) biofam.seq <- seqdef(biofam[501:600,10:25]) ## Indel and substitution costs based on log of inverse state frequencies lifcost <- seqcost(biofam.seq, method="INDELSLOG") ## Here lifcost$indel is a vector biofam.om <- seqdist(biofam.seq, method="OM", indel=lifcost$indel, sm=lifcost$sm) ## Optimal matching using transition rates based substitution-cost matrix ## and the associated indel cost ## Here trcost$indel is a scalar trcost <- seqcost(biofam.seq, method="TRATE") biofam.om <- seqdist(biofam.seq, method="OM", indel=trcost$indel, sm=trcost$sm) ## Using costs based on FUTURE with a forward lag of 4 fucost <- seqcost(biofam.seq, method="FUTURE", lag=4) biofam.om <- seqdist(biofam.seq, method="OM", indel=fucost$indel, sm=fucost$sm) ## Optimal matching using a unique substitution cost of 2 ## and an insertion/deletion cost of 3 ccost <- seqsubm(biofam.seq, method="CONSTANT", cval=2) biofam.om.c2 <- seqdist(biofam.seq, method="OM",indel=3, sm=ccost) ## Displaying the distance matrix for the first 10 sequences biofam.om.c2[1:10,1:10] ## ================================= ## Example with weights and missings ## ================================= data(ex1) ex1.seq <- seqdef(ex1[,1:13], weights=ex1$weights) ## Unweighted subm <- seqcost(ex1.seq, method="INDELSLOG", with.missing=TRUE, weighted=FALSE) ex1.om <- seqdist(ex1.seq, method="OM", indel=subm$indel, sm=subm$sm, with.missing=TRUE) ## Weighted subm.w <- seqcost(ex1.seq, method="INDELSLOG", with.missing=TRUE, weighted=TRUE) ex1.omw <- seqdist(ex1.seq, method="OM", indel=subm.w$indel, sm=subm.w$sm, with.missing=TRUE) ex1.om == ex1.omw